|
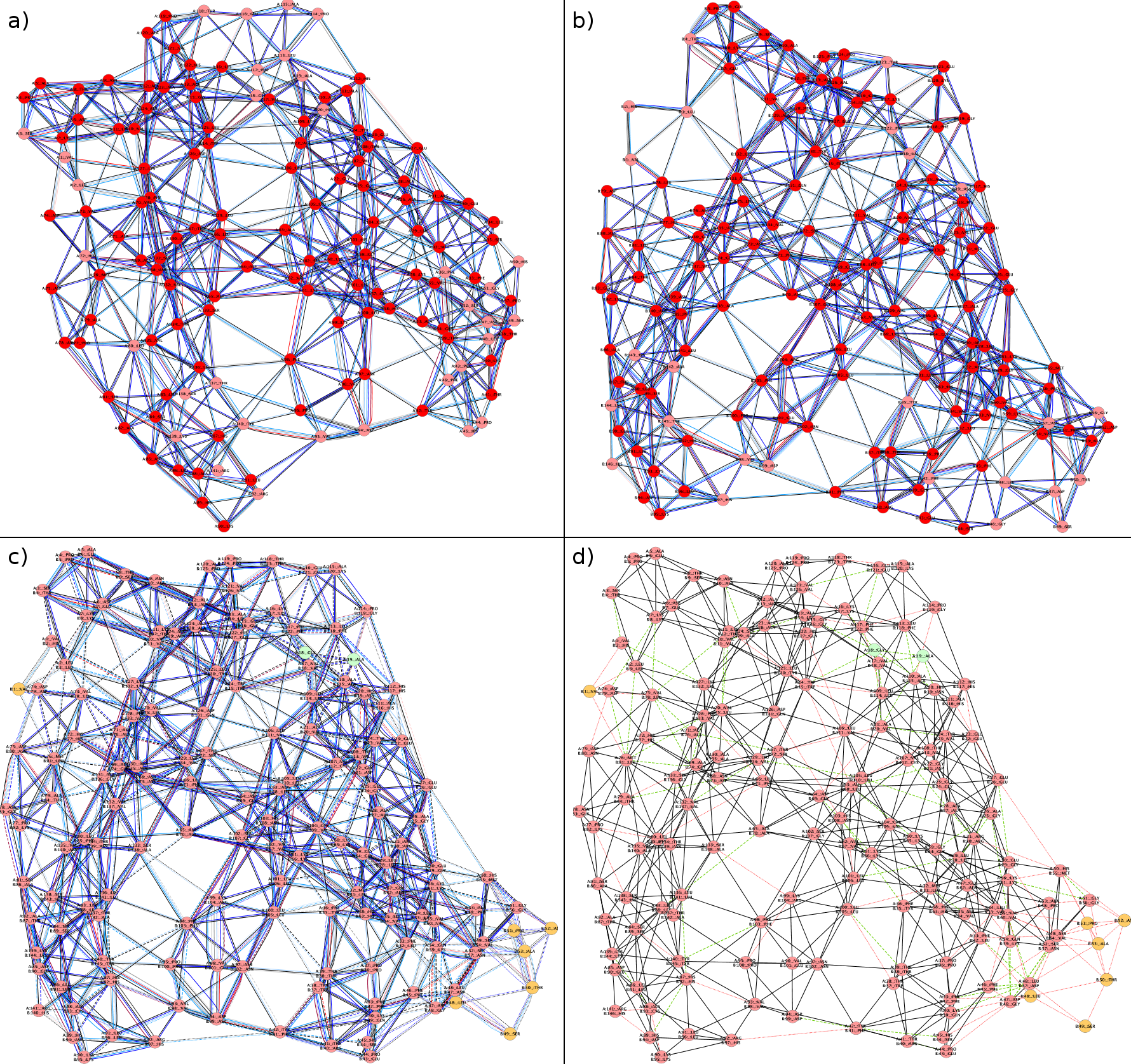
Example 4: RIN comparison of the alpha and beta chain of Deoxyhemoglobin based on an alignment of the corresponding 3D protein structures (click on the image to enlarge it)
- RIN representing the alpha chain (PDB identifier 4HHB) 1
- RIN representing the beta chain (PDB identifier 4HHB) 1
- Combined RIN with different types of non-covalent interactions 2,3,4
- Combined RIN with generic residue interactions 3,5
1Nodes colored according to secondary structure (red for helices, blue for strands).
2Nodes present only in the alpha chain are green, nodes present only in the beta chain are orange, and all other nodes are pink.
3Solid edge lines represent non-covalent residue interactions preserved in both structures; dashed interactions are present only in the alpha chain and dotted interactions only in the beta chain.
4Edges are colored according to interaction type (interatomic contacts in blue, hydrogen bonds in red, overlaps in gray).
5Interactions between residues only in the alpha chain are green, interactions only in the beta chain are red, and all other interaction edges are black.
|
 |